How We Measure
We currently report two metrics:
- R (relative) portion of YCR. This is the RCR recalculated strictly from open data. More details appear below.
- PaperRank. Computed with formulas from two papers:
- Recent advances in bibliometric indexes and the PaperRank problem (2014) by Pierluigi Amodio and Luigi Brugnano.
- Implementation of the PaperRank and AuthorRank indices in the Scopus database (2021) by Pierluigi Amodio, Luigi Brugnano, and Filippo Scarselli.
- Method A: original procedure with the default weight of 1.
- Method B: same procedure, but the default weight is 1 + R, where R is the relative portion of YCR. This adjustment gives the input signal a more realistic initial value.
How We Measure the R (relative) Portion of YCR
The R (relative) portion of YCR is currently calculated by an independent implementation of the
RCR algorithm. This metric was originally designed by the team of B. Ian Hutchins, Xin Yuan,
James M. Anderson, and George M. Santangelo in 2016 to evaluate relative performance of scientific
publications. More about RCR is here.
Important changes in our implementation:
- Calculations of IFs (impact factors) of journals are based on open data, which means transparency. Original RCR relies on imported IFs from a commercial provider.
- Calculations take into account all relevant cross-linked articles from open data. Original RCR relies on PubMed indexed articles only.
Overview of algorithms and methods to calculate RCR for the combined Oncology and Biomedical fields, based on the OpenAlex dataset:
- How we define our cluster of the Oncology field papers
- How we define our cluster of the Biomedical field papers
- How we calculate RCR metric based on OA dataset
How we define our cluster of Oncology field papers
We prepared a list of a total 588 onco-terms (separate words and phrases) by following methods:
- By using our common sense and some medical background of our team members
- By asking an LLM AIs, then carefully checking these suggestions manually
- By extracting all articles keywords from the cluster defined by onco-topics in OpenAlex (OA) dataset, more about this below
- By final combining, manual checking and cleaning
Then we took in all the articles from the OpenAlex (OA) dataset which contain any of these onco-terms
in title or in abstract or in keywords (generated by OA).
Additionally we took in all the topics from OA which contain any of these onco-terms in topics fields:
display_name, description, keywords. Then we manually checked all these topics and prepared a final
list of 412 topics that belong to the Oncology field.
Using a prepared list of onco-topics we took in all the articles which contain one of these topics
as primary topic (primerness was calculated by OpenAlex and is shown in their dataset).
So finally we define our cluster of Oncology field articles (18,085,165) by any of 2 rules:
- Article should contain any of onco-terms in title or in abstract or in keywords
- OR article should have primary topic from our list of onco-topics
How we define our cluster of Biomedical field papers
We prepared a list of topics, associated with the biomedical science by selecting OpenAlex topics whose "field" or "subfield" parameters indicate biomedical relevance. We chose 11 "fields", and 9 "subfields", related to biomedical science. Then we took in all the articles which contain one of these topics as primary topic, or as regular topic with score > 0.8. This way we define our cluster of Biomedical field articles (83,922,018).
How we calculate RCR metric based on OA dataset
To calculate the RCR metric we used the method described in the article
https://doi.org/10.1371/journal.pbio.1002541
by authors of RCR: B. Ian Hutchins, Xin Yuan, James M. Anderson, George M. Santangelo.
To understand code and algorithms we studied scripts from their github repository
https://github.com/NIHOPA/Relative...
.
First of all we calculate impact factors (IFs) for all the journals of OA.
For that we use the formula of classic 2-year impact factor:
the number of citations of articles published in 2 previous years and made in the IF year, divided by the total number of articles published in the 2 previous years.
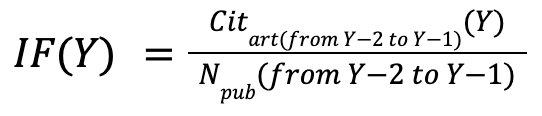
We used OpenAlex data about citations of articles and the primary journal of each article,
so we can count all the articles of each journal for a specific year and all their citations.
We used journals only (OpenAlex sources with type “journal”, not “book” and others) here.
We used articles only (OpenAlex work with type “article”, not “book” and others) here.
Then we define 1,444,476 “etalon” articles for all years from 1970 till 2023. For that we used the NIH RePORTER system and took in only articles (not books etc) made under Research Grant with activity code R01 which matched the initial description of etalons in RCR paper.
For each article of these etalons we define a co-citation network: we took articles which
cite one initial etalon and then took all of their references.
Then for each paper of the co-citation network we took a precalculated OA-based impact factor
of its primary journal for the year of the paper's publication. After that we define
Field Citation Rate (FCR) of each etalon work by taking an average of all IFs of articles
from a co-citation network.
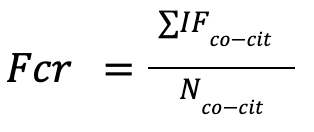
Then for each etalon article we define the metric Citations Per Year (CPY) using the number of its citations from year of publication till this year (2024) divided by number of years from year of publication till this year (2024).
![]()
Then we split all of etalon articles by year of publication and for each year we calculate Quantile Regression coefficients (QR coeffs) using FCR and CPY. For that we used a function “quantreg” from python library statsmodel and assuming:
![]()
So for each year we define 2 coefficients: b and a and we will use them to calculate
RCR for non-etalon articles of corresponding years.
Then to calculate RCR for any non-etalon article we made pretty similar steps:
- define co-citation network of article
- calculate CPY of article and FCR of its co-citation network using OA-based IFs
- calculate Expected Citations Rate (ECR) using QR coeffs (based on etalons) of publication year and article FCR:
- calculate RCR for article using CPY and ECR:
![]()
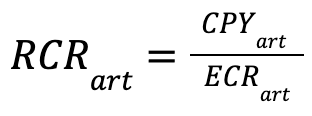
This way we calculate classic RCR for all articles (etalons and non-etalons) from our combined clusters of the Oncology and Biomedical field papers and save their values to our database. As confirmation of the correctness of our algorithm implementation, we verified that the median RCR value calculated for all etalon articles is approximately equal to 1.
If you look for the RCR value of an article outside of our combined clusters of the Oncology and Biomedical field papers then keep in mind that Biomed R01 etalons were used for non-biomed articles to calculate their RCR values.